Difference Between Whole Genome Sequencing and Next Generation Sequencing
• Categorized under Science,Technology | Difference Between Whole Genome Sequencing and Next Generation Sequencing
In the past there decades, our understanding of the human genome has increased exponentially, together with the rapid growth of sequence databases and bioinformatics tools. And the pressure to produce more, faster, and cheaper DNA sequences become pivotal for scientific research. In the first decade of the twenty-first century, advances in microarray techniques allowed scientists and researchers to analyze the human genome with increasing resolution. The conventional techniques have their limitations that call for the development of new experimental methods. The Sanger sequencing method which is still considered as the gold standard for sequencing has its limitations. With the ability to sequence more than a million DNA fragments at a time, the next-generation sequencing (NGS) has revolutionized the ability to generate large volumes of sequence data at an extremely low cost.
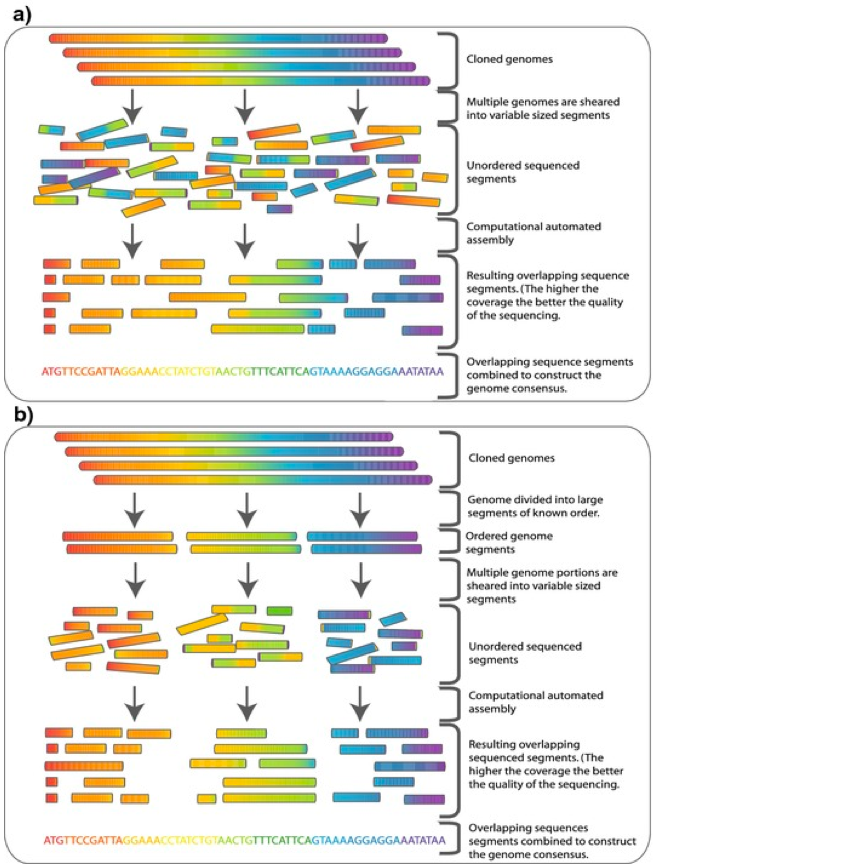
What is Whole Genome Sequencing?
Whole genome sequencing (WGS) is a comprehensive method of analyzing the entire genomic DNA sequence of a cell at a single time. WGS is a technology that enables scientists to read the exact sequence of all the letters that make up your complete set of DNA. The human genome is still poorly understood and even the best studied portion of the genome is represented by approximately 3% of the genome that encodes proteins. And the functions of the proteins in the human genome are also partially understood. For these reasons, the computational analysis of genome sequence data aims to indentify the medical significance of all the functional elements of the genome and identify gene functions and their involvement in disease. WGS using NGS technologies is anticipated to revolutionize clinical care, yet the general promise of whole genome sequencing is far from being fulfilled. WGS involves sequencing the whole genome to study mutations and rearrangements.
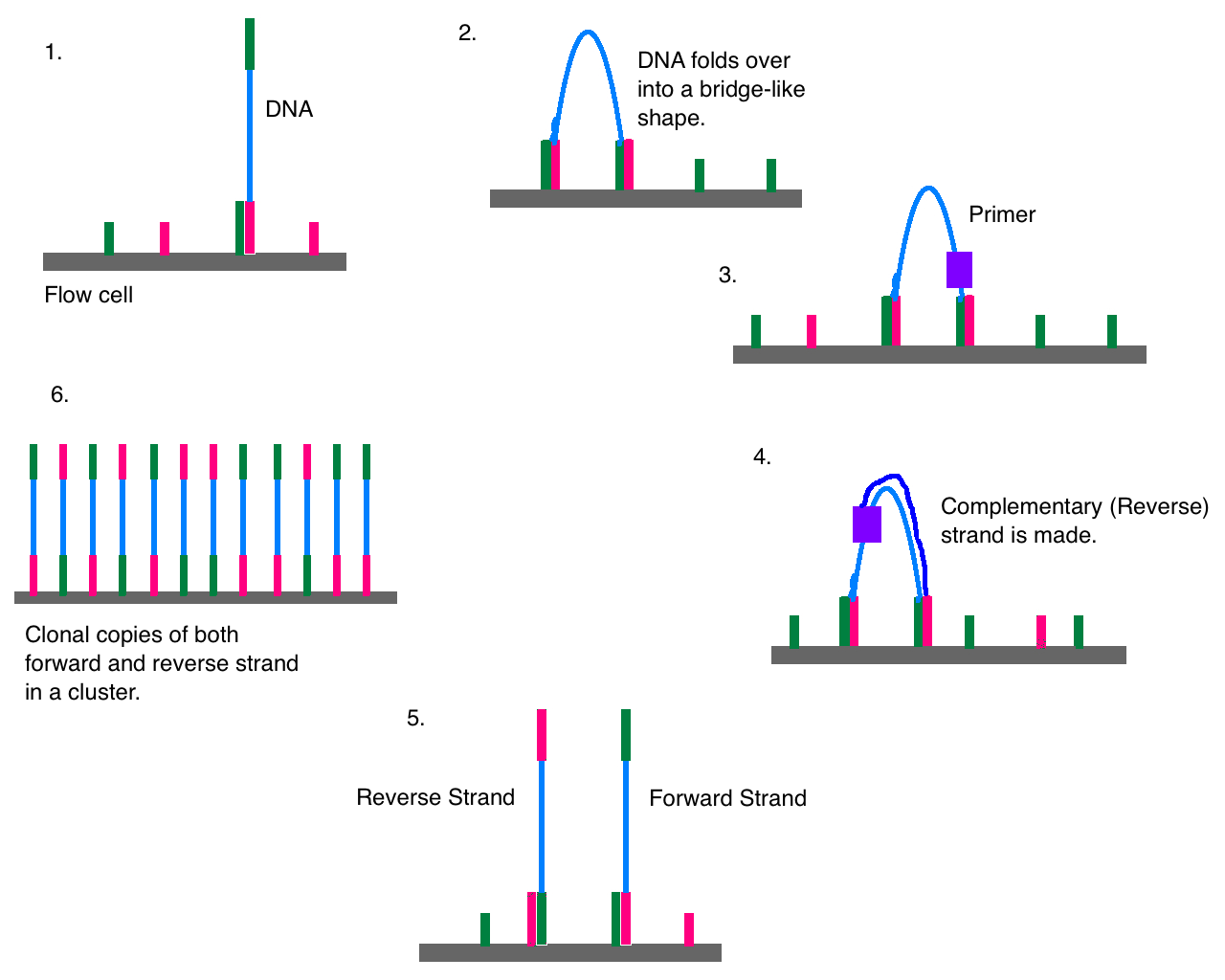
What is Next Generation Sequencing?
There is no precise definition of next generation sequencing (NGS), but there are several features that clearly distinguish NGS platforms from conventional capillary-based sequencing. Next generation sequencing, also known as massively parallel sequencing is a relatively new technique that allows the sequencing of a DNA molecule with total size significantly larger than 1 million base pairs in a single experiment. NGS is a powerful platform that has revolutionized the field of biological sciences by demonstrating the capacity to DNA sequencing at unprecedented speed and offering a high throughput option, allowing sequencing of multiple individuals at the same time. This is achieved by miniaturizing the volume of individual sequencing reactions, which limits the size of the instruments and minimizes the cost of reagents per reaction. NGS is applied in a variety of areas including de novo genome sequencing, whole genome resequencing, transcriptome analysis, targeted resequencing, small RNA and miRNA sequencing, whole exome sequencing, etc.
Difference between Whole Genome Sequencing and Next Generation Sequencing
Technology
– Whole genome sequencing (WGS) is a comprehensive method of analyzing the entire genomic DNA sequence of a cell at a single time. The technology enables scientists to read the exact sequence of all the letters that make up your complete set of DNA. Next generation sequencing, also known as massively parallel sequencing is a relatively new technique that allows the sequencing of a DNA molecule with total size significantly larger than 1 million base pairs in a single experiment. The idea is to generate large amounts of DNA sequence data through massive parallelization.
Process
– WGS is a lab procedure that identifies the order of bases in the genome in a single process. In order to do this, a blood or saliva sample is taken and sent to the lab where a machine breaks up the DNA and reads the letters like a code. Researchers then compare your DNA sequence to a standardized reference and identify the variations between the two set of letters which lead to many different types of personal information being identified. The principle of NGS is similar to the traditional capillary electrophoresis method, which involves three basic steps: DNA fragmentation, sequencing the libraries, and data analysis.
Applications
– Whole genome sequencing involves sequencing the whole genome to study mutations and rearrangements. With whole genome sequencing, you can assemble genomes de novo, compare the genome of an organism to a reference genome, identify novel genome assembly, track pathogen outbreaks, molecular evolution, diagnosis of suspected Mendelian syndromes and cancer, and so much more. NGS technologies are used for whole genome sequencing, epigenetics, transcriptome analysis, de novo genome sequencing, large-scale analysis of DNA methylation, discovery of non-coding RNAs and protein-binding sites, biomarkers identification for early diagnosis, and more.
Next Generation Sequencing vs. Whole Genome Sequencing: Comparison Chart
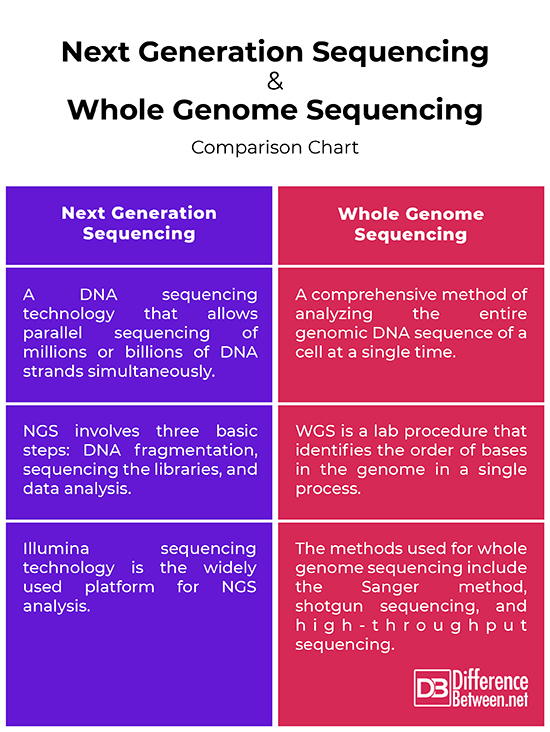
Summary of Next Generation Sequencing vs. Whole Genome Sequencing
There is no clear definition of next generation sequencing (NGS), but there are several features that clearly distinguish NGS platforms from conventional DNA sequencing methods such as the already-popular Sanger method, which is still considered by many as the gold standard of DNA sequencing. NGS technologies are routinely applied in contemporary molecular research and have been a powerful tool for many clinical applications. It is a great diagnostic platform as it provides the potential for comprehensive mutation detection over multiple genomic targets at a time, high-throughput analysis of patient panels across disease loci, and discovery of non-coding RNAs and protein-binding sites.
Outside his professional life, Sagar loves to connect with people from different cultures and origin. You can say he is curious by nature. He believes everyone is a learning experience and it brings a certain excitement, kind of a curiosity to keep going. It may feel silly at first, but it loosens you up after a while and makes it easier for you to start conversations with total strangers – that’s what he said."
- Difference Between Caucus and Primary - June 18, 2024
- Difference Between PPO and POS - May 30, 2024
- Difference Between RFID and NFC - May 28, 2024
Sharing is caring!
Search DifferenceBetween.net :
Cite
APA 7
Khillar, S. (2021, November 23). Difference Between Whole Genome Sequencing and Next Generation Sequencing. Difference Between Similar Terms and Objects. http://www.differencebetween.net/technology/difference-between-whole-genome-sequencing-and-next-generation-sequencing/.
MLA 8
Khillar, Sagar. "Difference Between Whole Genome Sequencing and Next Generation Sequencing." Difference Between Similar Terms and Objects, 23 November, 2021, http://www.differencebetween.net/technology/difference-between-whole-genome-sequencing-and-next-generation-sequencing/.
Leave a Response
Written by : Sagar Khillar. and updated on 2021, November 23
References :
[0]Demkow, Urszula and Rafal Ploski. Clinical Applications for Next-Generation Sequencing. Massachusetts, United States: Academic Press, 2015. Print
[1]Liu, Zhanjiang (John). Next Generation Sequencing and Whole Genome Selection in Aquaculture. New Jersey, United States: John Wiley & Sons, 2010. Print
[2]Janitz, Michal. Next-Generation Genome Sequencing: Towards Personalized Medicine. New Jersey, United States: John Wiley & Sons, 2010. Print
[3]Kulkarni, Shashikant and John Pfeifer. Clinical Genomics. Massachusetts, United States: Academic Press, 2014. Print
[4]Robinson, Peter N. et al. Computational Exome and Genome Analysis. Florida, United States: CRC Press, 2017. Print
[5]Image credit: https://commons.wikimedia.org/wiki/File:Cluster_Generation.png
[6]Image credit: https://commons.wikimedia.org/wiki/File:Whole_genome_shotgun_sequencing_versus_Hierarchical_shotgun_sequencing.png
See more about : Next Generation Sequencing, Whole Genome Sequencing
